XClone
XClone is implementated on Python 3. XClone integrates expression levels and allelic balance to detect haplotype-aware CNVs and reconstruct tumour clonal substructure from scRNA-seq data.
XClone algorithm
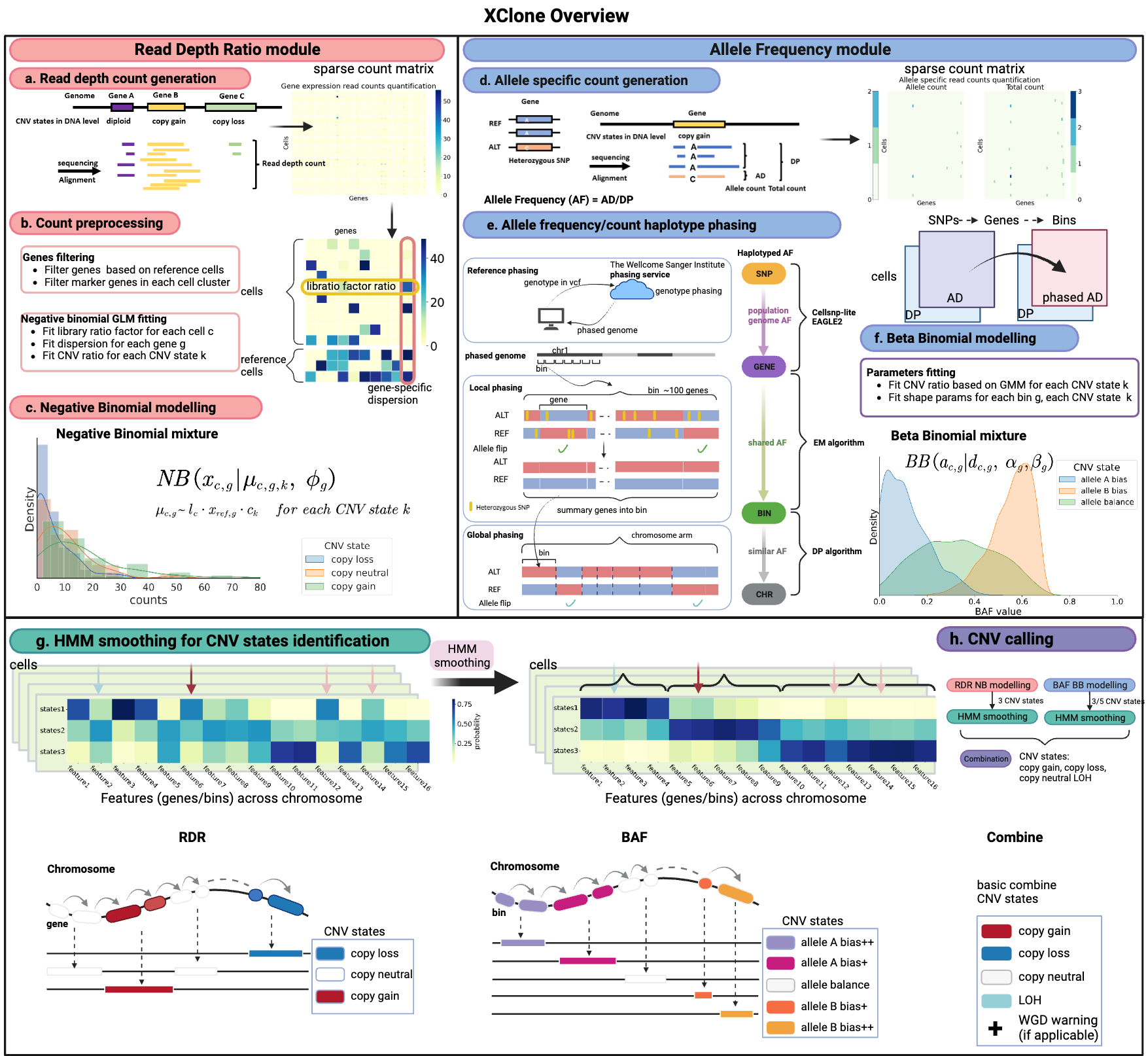
XClone has two modules of information: the read depth ratio (RDR) module and the B-allele frequency (BAF) module, where each of the modules has its own CNV states and noise models for likelihood function.
XClone RDR module
In the RDR module, XClone considers three CNV states about the absolute copy numbers: copy loss, copy neutral and copy gain. It takes the raw read or UMI counts as input and models the noise via a negative-binomial distribution.
XClone BAF module
In the BAF module, we introduce a three-step phasing strategy to aggregate allelic features:
from one SNP to multiple SNPs on a gene
from a single gene to a mega gene
from a mega gene to a whole chromosome arm
XClone takes the phased mega gene as the feature and defines three allele-based CNV states: allele A drop, allele balance and allele B drop. By taking the allelic count matrices of both alleles, it models the read or UMI counts of the B allele for each feature in each cell via a beta-binomial distribution.