XClone: detection of allele-specific subclonal copy number variations from single-cell transcriptomic data
XClone’s key features
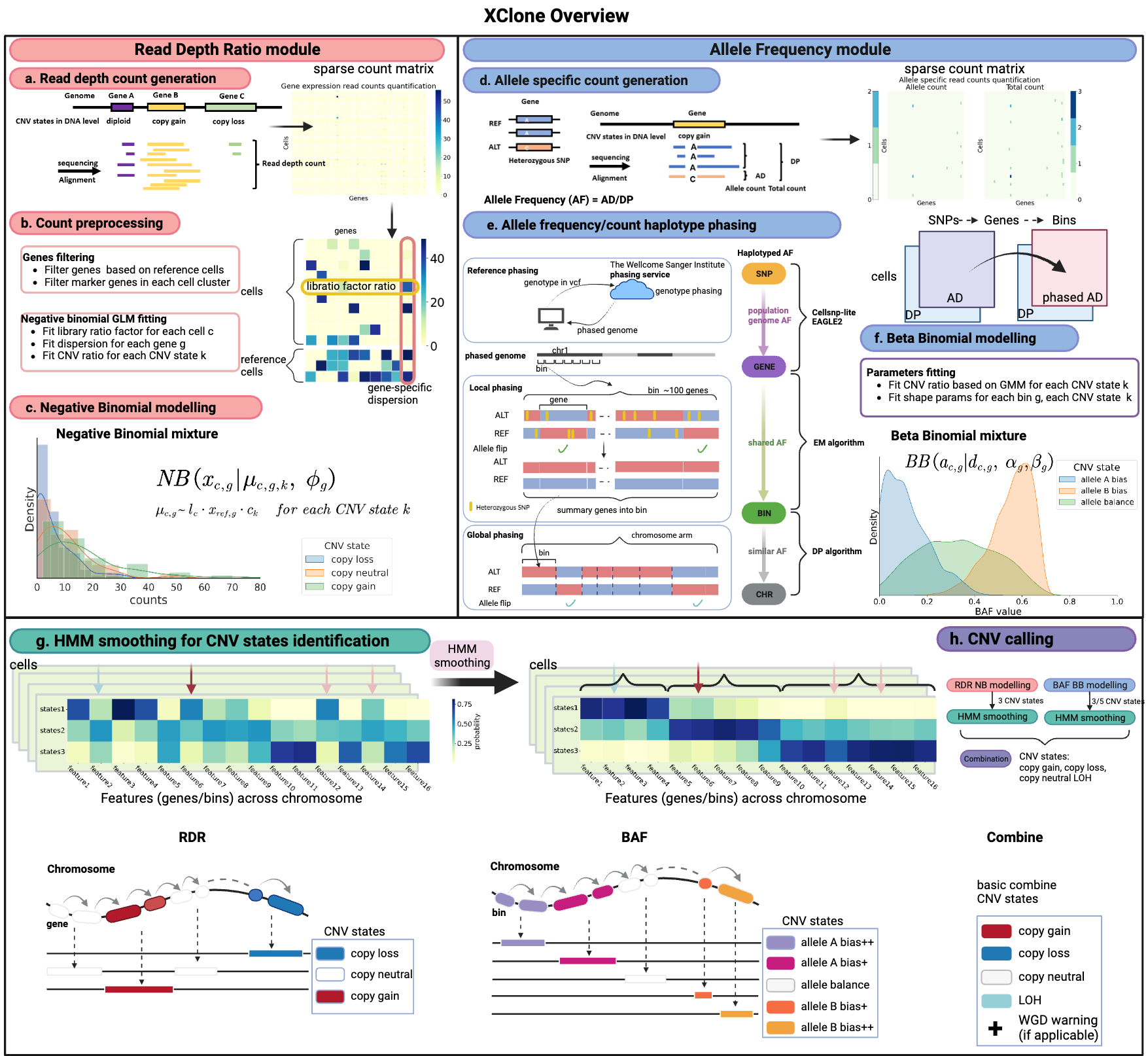
XClone has two modules of information: the read depth ratio (RDR) module and the B-allele frequency (BAF) module, where each of the modules has its own CNV states and noise models for likelihood function.
XClone introduces two orthogonal methods for smoothing the probabilities of assigning CNV states for each feature in each cell.
XClone introduces a post-step for combining the CNV states computed from the RDR and BAF modules.
Latest news
References
For details of the method, please checkout our paper [1].
Support
Found a bug or would like to see a feature implemented? Feel free to submit an issue. Have a question or would like to start a new discussion? Head over to GitHub discussions. In either case, you can also always send us an email. Your help to improve XClone is highly appreciated. For further information visit xclone.org.
